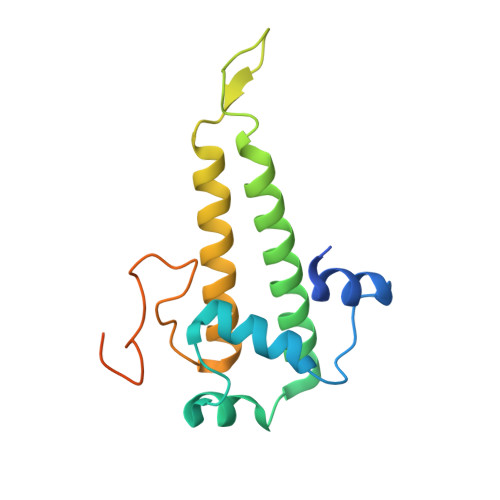
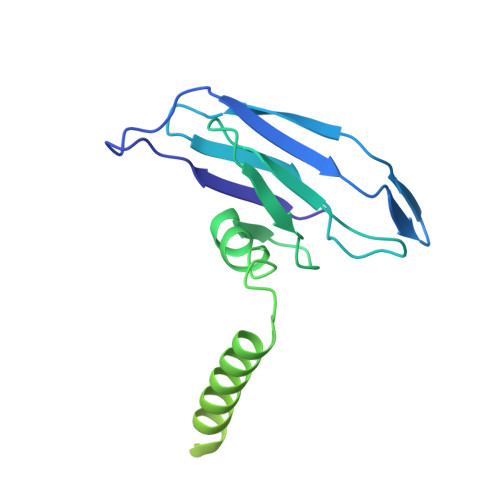
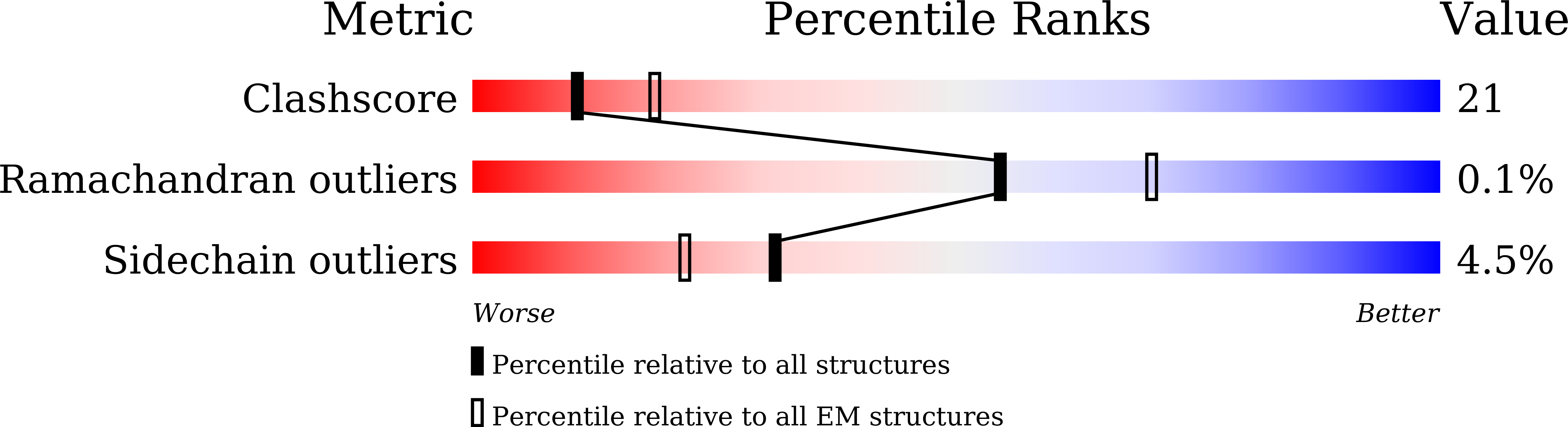Structure of the intact tail machine of Anabaena myophage A-1(L).
Yu, R.C., Yang, F., Zhang, H.Y., Hou, P., Du, K., Zhu, J., Cui, N., Xu, X., Chen, Y., Li, Q., Zhou, C.Z.(2024) Nat Commun 15: 2654-2654
- PubMed: 38531972
- DOI: https://doi.org/10.1038/s41467-024-47006-z
- Primary Citation of Related Structures:
8KE9, 8KEA, 8KEC, 8KEE, 8KEF, 8KEG, 8TS6 - PubMed Abstract:
The Myoviridae cyanophage A-1(L) specifically infects the model cyanobacteria Anabaena sp. PCC 7120. Following our recent report on the capsid structure of A-1(L), here we present the high-resolution cryo-EM structure of its intact tail machine including the neck, tail and attached fibers. Besides the dodecameric portal, the neck contains a canonical hexamer connected to a unique pentadecamer that anchors five extended bead-chain-like neck fibers. The 1045-Å-long contractile tail is composed of a helical bundle of tape measure proteins surrounded by a layer of tube proteins and a layer of sheath proteins, ended with a five-component baseplate. The six long and six short tail fibers are folded back pairwise, each with one end anchoring to the baseplate and the distal end pointing to the capsid. Structural analysis combined with biochemical assays further enable us to identify the dual hydrolytic activities of the baseplate hub, in addition to two host receptor binding domains in the tail fibers. Moreover, the structure of the intact A-1(L) also helps us to reannotate its genome. These findings will facilitate the application of A-1(L) as a chassis cyanophage in synthetic biology.
Organizational Affiliation:
School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, China.